데이터 준비 🌱
BioProject : PRJNA1156939
- 30명의 소아 크론병 환자(치료 전)의 대변, 대장 및 말단 회장 샘플 및 비슷한 연령의 건강한 대조군 36명의 대변 샘플 (16S rRNA의 V3-V4)
| Source | Case | Count |
| Stool | Patient | 30 |
| Stool | Healthy | 36 |
| Colon | Patient | 30 |
| Terminal ileum | Patient | 30 |
1) Fastq 파일 다운로드 받기
ENA Broswer에서 Download report을 클릭하여 해당 프로젝트의 메타데이터를 다운로드 받을 수 있으며, 다운로드 받은 TSV 파일에는 각 샘플별로 ftp 주소를 확인 할 수 있습니다.
ENA Broswer에서 fastq_ftp 주소 얻는 방법

# download.sh
mkdir -p /root/qiime2/input
cd /root/qiime2/input
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/000/SRR30554300/SRR30554300_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/000/SRR30554300/SRR30554300_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/001/SRR30554301/SRR30554301_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/001/SRR30554301/SRR30554301_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/002/SRR30554302/SRR30554302_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/002/SRR30554302/SRR30554302_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/003/SRR30554303/SRR30554303_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/003/SRR30554303/SRR30554303_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/004/SRR30554304/SRR30554304_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/004/SRR30554304/SRR30554304_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/005/SRR30554305/SRR30554305_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/005/SRR30554305/SRR30554305_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/006/SRR30554306/SRR30554306_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/006/SRR30554306/SRR30554306_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/007/SRR30554307/SRR30554307_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/007/SRR30554307/SRR30554307_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/008/SRR30554308/SRR30554308_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/008/SRR30554308/SRR30554308_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/009/SRR30554309/SRR30554309_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/009/SRR30554309/SRR30554309_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/010/SRR30554310/SRR30554310_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/010/SRR30554310/SRR30554310_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/011/SRR30554311/SRR30554311_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/011/SRR30554311/SRR30554311_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/012/SRR30554312/SRR30554312_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/012/SRR30554312/SRR30554312_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/013/SRR30554313/SRR30554313_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/013/SRR30554313/SRR30554313_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/014/SRR30554314/SRR30554314_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/014/SRR30554314/SRR30554314_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/015/SRR30554315/SRR30554315_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/015/SRR30554315/SRR30554315_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/016/SRR30554316/SRR30554316_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/016/SRR30554316/SRR30554316_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/017/SRR30554317/SRR30554317_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/017/SRR30554317/SRR30554317_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/018/SRR30554318/SRR30554318_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/018/SRR30554318/SRR30554318_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/019/SRR30554319/SRR30554319_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/019/SRR30554319/SRR30554319_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/020/SRR30554320/SRR30554320_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/020/SRR30554320/SRR30554320_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/021/SRR30554321/SRR30554321_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/021/SRR30554321/SRR30554321_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/022/SRR30554322/SRR30554322_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/022/SRR30554322/SRR30554322_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/023/SRR30554323/SRR30554323_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/023/SRR30554323/SRR30554323_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/024/SRR30554324/SRR30554324_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/024/SRR30554324/SRR30554324_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/025/SRR30554325/SRR30554325_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/025/SRR30554325/SRR30554325_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/026/SRR30554326/SRR30554326_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/026/SRR30554326/SRR30554326_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/027/SRR30554327/SRR30554327_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/027/SRR30554327/SRR30554327_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/028/SRR30554328/SRR30554328_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/028/SRR30554328/SRR30554328_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/029/SRR30554329/SRR30554329_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/029/SRR30554329/SRR30554329_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/030/SRR30554330/SRR30554330_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/030/SRR30554330/SRR30554330_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/031/SRR30554331/SRR30554331_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/031/SRR30554331/SRR30554331_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/032/SRR30554332/SRR30554332_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/032/SRR30554332/SRR30554332_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/033/SRR30554333/SRR30554333_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/033/SRR30554333/SRR30554333_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/034/SRR30554334/SRR30554334_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/034/SRR30554334/SRR30554334_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/035/SRR30554335/SRR30554335_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/035/SRR30554335/SRR30554335_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/036/SRR30554336/SRR30554336_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/036/SRR30554336/SRR30554336_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/037/SRR30554337/SRR30554337_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/037/SRR30554337/SRR30554337_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/038/SRR30554338/SRR30554338_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/038/SRR30554338/SRR30554338_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/039/SRR30554339/SRR30554339_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/039/SRR30554339/SRR30554339_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/040/SRR30554340/SRR30554340_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/040/SRR30554340/SRR30554340_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/041/SRR30554341/SRR30554341_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/041/SRR30554341/SRR30554341_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/042/SRR30554342/SRR30554342_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/042/SRR30554342/SRR30554342_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/043/SRR30554343/SRR30554343_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/043/SRR30554343/SRR30554343_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/044/SRR30554344/SRR30554344_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/044/SRR30554344/SRR30554344_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/045/SRR30554345/SRR30554345_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/045/SRR30554345/SRR30554345_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/046/SRR30554346/SRR30554346_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/046/SRR30554346/SRR30554346_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/047/SRR30554347/SRR30554347_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/047/SRR30554347/SRR30554347_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/048/SRR30554348/SRR30554348_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/048/SRR30554348/SRR30554348_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/049/SRR30554349/SRR30554349_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/049/SRR30554349/SRR30554349_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/050/SRR30554350/SRR30554350_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/050/SRR30554350/SRR30554350_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/051/SRR30554351/SRR30554351_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/051/SRR30554351/SRR30554351_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/052/SRR30554352/SRR30554352_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/052/SRR30554352/SRR30554352_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/053/SRR30554353/SRR30554353_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/053/SRR30554353/SRR30554353_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/054/SRR30554354/SRR30554354_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/054/SRR30554354/SRR30554354_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/055/SRR30554355/SRR30554355_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/055/SRR30554355/SRR30554355_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/056/SRR30554356/SRR30554356_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/056/SRR30554356/SRR30554356_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/057/SRR30554357/SRR30554357_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/057/SRR30554357/SRR30554357_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/058/SRR30554358/SRR30554358_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/058/SRR30554358/SRR30554358_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/059/SRR30554359/SRR30554359_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/059/SRR30554359/SRR30554359_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/060/SRR30554360/SRR30554360_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/060/SRR30554360/SRR30554360_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/061/SRR30554361/SRR30554361_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/061/SRR30554361/SRR30554361_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/062/SRR30554362/SRR30554362_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/062/SRR30554362/SRR30554362_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/063/SRR30554263/SRR30554263_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/063/SRR30554263/SRR30554263_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/063/SRR30554363/SRR30554363_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/063/SRR30554363/SRR30554363_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/064/SRR30554264/SRR30554264_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/064/SRR30554264/SRR30554264_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/064/SRR30554364/SRR30554364_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/064/SRR30554364/SRR30554364_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/065/SRR30554265/SRR30554265_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/065/SRR30554265/SRR30554265_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/065/SRR30554365/SRR30554365_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/065/SRR30554365/SRR30554365_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/066/SRR30554266/SRR30554266_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/066/SRR30554266/SRR30554266_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/066/SRR30554366/SRR30554366_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/066/SRR30554366/SRR30554366_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/067/SRR30554267/SRR30554267_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/067/SRR30554267/SRR30554267_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/067/SRR30554367/SRR30554367_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/067/SRR30554367/SRR30554367_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/068/SRR30554268/SRR30554268_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/068/SRR30554268/SRR30554268_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/068/SRR30554368/SRR30554368_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/068/SRR30554368/SRR30554368_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/069/SRR30554269/SRR30554269_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/069/SRR30554269/SRR30554269_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/069/SRR30554369/SRR30554369_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/069/SRR30554369/SRR30554369_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/070/SRR30554270/SRR30554270_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/070/SRR30554270/SRR30554270_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/070/SRR30554370/SRR30554370_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/070/SRR30554370/SRR30554370_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/071/SRR30554271/SRR30554271_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/071/SRR30554271/SRR30554271_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/071/SRR30554371/SRR30554371_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/071/SRR30554371/SRR30554371_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/072/SRR30554272/SRR30554272_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/072/SRR30554272/SRR30554272_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/072/SRR30554372/SRR30554372_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/072/SRR30554372/SRR30554372_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/073/SRR30554273/SRR30554273_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/073/SRR30554273/SRR30554273_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/073/SRR30554373/SRR30554373_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/073/SRR30554373/SRR30554373_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/074/SRR30554274/SRR30554274_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/074/SRR30554274/SRR30554274_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/074/SRR30554374/SRR30554374_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/074/SRR30554374/SRR30554374_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/075/SRR30554275/SRR30554275_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/075/SRR30554275/SRR30554275_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/075/SRR30554375/SRR30554375_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/075/SRR30554375/SRR30554375_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/076/SRR30554276/SRR30554276_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/076/SRR30554276/SRR30554276_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/076/SRR30554376/SRR30554376_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/076/SRR30554376/SRR30554376_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/077/SRR30554277/SRR30554277_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/077/SRR30554277/SRR30554277_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/077/SRR30554377/SRR30554377_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/077/SRR30554377/SRR30554377_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/078/SRR30554278/SRR30554278_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/078/SRR30554278/SRR30554278_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/078/SRR30554378/SRR30554378_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/078/SRR30554378/SRR30554378_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/079/SRR30554279/SRR30554279_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/079/SRR30554279/SRR30554279_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/079/SRR30554379/SRR30554379_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/079/SRR30554379/SRR30554379_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/080/SRR30554280/SRR30554280_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/080/SRR30554280/SRR30554280_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/080/SRR30554380/SRR30554380_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/080/SRR30554380/SRR30554380_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/081/SRR30554281/SRR30554281_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/081/SRR30554281/SRR30554281_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/081/SRR30554381/SRR30554381_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/081/SRR30554381/SRR30554381_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/082/SRR30554282/SRR30554282_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/082/SRR30554282/SRR30554282_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/082/SRR30554382/SRR30554382_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/082/SRR30554382/SRR30554382_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/083/SRR30554283/SRR30554283_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/083/SRR30554283/SRR30554283_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/083/SRR30554383/SRR30554383_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/083/SRR30554383/SRR30554383_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/084/SRR30554284/SRR30554284_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/084/SRR30554284/SRR30554284_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/084/SRR30554384/SRR30554384_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/084/SRR30554384/SRR30554384_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/085/SRR30554285/SRR30554285_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/085/SRR30554285/SRR30554285_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/085/SRR30554385/SRR30554385_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/085/SRR30554385/SRR30554385_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/086/SRR30554286/SRR30554286_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/086/SRR30554286/SRR30554286_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/086/SRR30554386/SRR30554386_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/086/SRR30554386/SRR30554386_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/087/SRR30554287/SRR30554287_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/087/SRR30554287/SRR30554287_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/087/SRR30554387/SRR30554387_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/087/SRR30554387/SRR30554387_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/088/SRR30554288/SRR30554288_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/088/SRR30554288/SRR30554288_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/088/SRR30554388/SRR30554388_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/088/SRR30554388/SRR30554388_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/089/SRR30554289/SRR30554289_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/089/SRR30554289/SRR30554289_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/090/SRR30554290/SRR30554290_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/090/SRR30554290/SRR30554290_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/091/SRR30554291/SRR30554291_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/091/SRR30554291/SRR30554291_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/092/SRR30554292/SRR30554292_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/092/SRR30554292/SRR30554292_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/093/SRR30554293/SRR30554293_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/093/SRR30554293/SRR30554293_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/094/SRR30554294/SRR30554294_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/094/SRR30554294/SRR30554294_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/095/SRR30554295/SRR30554295_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/095/SRR30554295/SRR30554295_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/096/SRR30554296/SRR30554296_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/096/SRR30554296/SRR30554296_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/097/SRR30554297/SRR30554297_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/097/SRR30554297/SRR30554297_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/098/SRR30554298/SRR30554298_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/098/SRR30554298/SRR30554298_2.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/099/SRR30554299/SRR30554299_1.fastq.gz
wget ftp.sra.ebi.ac.uk/vol1/fastq/SRR305/099/SRR30554299/SRR30554299_2.fastq.gz
sh download.sh만약 $'\r': command not found 라는 오류가 발생했다면 아래의 포스팅을 참고해주세요.
🔗 2025.03.04 - [코딩/Linux] - [ERROR] $'\r': command not found
[ERROR] $'\r': command not found
❓원인$'\r': command not found에러는 windows에서 작성한 스크립트를 linux에서 실행할 때 발생하는 에러입니다.윈도우에서 작성한 스크립트에서 개행 시 자동으로 \r\n이 붙는데, linux에서는 인식할 수
jeonyeong.tistory.com
2) Manifest 파일 제작하기
Manifest 파일은 각 샘플에 대한 raw data(fastq) 파일의 절대 경로를 담고 있습니다. Paired-end 데이터를 사용할 경우, Manifest 파일 포멧은 아래와 같습니다.
| sample-id | forward-absolute-filepath | reverse-absolute-filepath |
| sample1 | sample1 forward 서열의 절대 경로 |
sample1 reverse 서열의 절대 경로 |
| sample2 | sample2 forward 서열의 절대 경로 |
sample2 reverse 서열의 절대 경로 |
저는 /root/qiime2/input이라는 경로에 모든 fastq 파일을 저장하였기 때문에, 아래와 같이 manifest 파일을 작성 할 수 있습니다. 이때, 열은 '탭'으로 구분합니다.
sample-id forward-absolute-filepath reverse-absolute-filepath
SRR30554265 /root/qiime2/input/SRR30554265_1.fastq.gz /root/qiime2/input/SRR30554265_2.fastq.gz
SRR30554270 /root/qiime2/input/SRR30554270_1.fastq.gz /root/qiime2/input/SRR30554270_2.fastq.gz
SRR30554271 /root/qiime2/input/SRR30554271_1.fastq.gz /root/qiime2/input/SRR30554271_2.fastq.gz
SRR30554273 /root/qiime2/input/SRR30554273_1.fastq.gz /root/qiime2/input/SRR30554273_2.fastq.gz
SRR30554277 /root/qiime2/input/SRR30554277_1.fastq.gz /root/qiime2/input/SRR30554277_2.fastq.gz
SRR30554264 /root/qiime2/input/SRR30554264_1.fastq.gz /root/qiime2/input/SRR30554264_2.fastq.gz
SRR30554266 /root/qiime2/input/SRR30554266_1.fastq.gz /root/qiime2/input/SRR30554266_2.fastq.gz
SRR30554278 /root/qiime2/input/SRR30554278_1.fastq.gz /root/qiime2/input/SRR30554278_2.fastq.gz
SRR30554267 /root/qiime2/input/SRR30554267_1.fastq.gz /root/qiime2/input/SRR30554267_2.fastq.gz
SRR30554269 /root/qiime2/input/SRR30554269_1.fastq.gz /root/qiime2/input/SRR30554269_2.fastq.gz
SRR30554279 /root/qiime2/input/SRR30554279_1.fastq.gz /root/qiime2/input/SRR30554279_2.fastq.gz
SRR30554272 /root/qiime2/input/SRR30554272_1.fastq.gz /root/qiime2/input/SRR30554272_2.fastq.gz
SRR30554280 /root/qiime2/input/SRR30554280_1.fastq.gz /root/qiime2/input/SRR30554280_2.fastq.gz
SRR30554281 /root/qiime2/input/SRR30554281_1.fastq.gz /root/qiime2/input/SRR30554281_2.fastq.gz
SRR30554282 /root/qiime2/input/SRR30554282_1.fastq.gz /root/qiime2/input/SRR30554282_2.fastq.gz
SRR30554283 /root/qiime2/input/SRR30554283_1.fastq.gz /root/qiime2/input/SRR30554283_2.fastq.gz
SRR30554292 /root/qiime2/input/SRR30554292_1.fastq.gz /root/qiime2/input/SRR30554292_2.fastq.gz
SRR30554285 /root/qiime2/input/SRR30554285_1.fastq.gz /root/qiime2/input/SRR30554285_2.fastq.gz
SRR30554286 /root/qiime2/input/SRR30554286_1.fastq.gz /root/qiime2/input/SRR30554286_2.fastq.gz
SRR30554293 /root/qiime2/input/SRR30554293_1.fastq.gz /root/qiime2/input/SRR30554293_2.fastq.gz
SRR30554288 /root/qiime2/input/SRR30554288_1.fastq.gz /root/qiime2/input/SRR30554288_2.fastq.gz
SRR30554296 /root/qiime2/input/SRR30554296_1.fastq.gz /root/qiime2/input/SRR30554296_2.fastq.gz
SRR30554295 /root/qiime2/input/SRR30554295_1.fastq.gz /root/qiime2/input/SRR30554295_2.fastq.gz
SRR30554297 /root/qiime2/input/SRR30554297_1.fastq.gz /root/qiime2/input/SRR30554297_2.fastq.gz
SRR30554300 /root/qiime2/input/SRR30554300_1.fastq.gz /root/qiime2/input/SRR30554300_2.fastq.gz
SRR30554299 /root/qiime2/input/SRR30554299_1.fastq.gz /root/qiime2/input/SRR30554299_2.fastq.gz
SRR30554303 /root/qiime2/input/SRR30554303_1.fastq.gz /root/qiime2/input/SRR30554303_2.fastq.gz
SRR30554304 /root/qiime2/input/SRR30554304_1.fastq.gz /root/qiime2/input/SRR30554304_2.fastq.gz
SRR30554309 /root/qiime2/input/SRR30554309_1.fastq.gz /root/qiime2/input/SRR30554309_2.fastq.gz
SRR30554306 /root/qiime2/input/SRR30554306_1.fastq.gz /root/qiime2/input/SRR30554306_2.fastq.gz
SRR30554307 /root/qiime2/input/SRR30554307_1.fastq.gz /root/qiime2/input/SRR30554307_2.fastq.gz
SRR30554311 /root/qiime2/input/SRR30554311_1.fastq.gz /root/qiime2/input/SRR30554311_2.fastq.gz
SRR30554308 /root/qiime2/input/SRR30554308_1.fastq.gz /root/qiime2/input/SRR30554308_2.fastq.gz
SRR30554313 /root/qiime2/input/SRR30554313_1.fastq.gz /root/qiime2/input/SRR30554313_2.fastq.gz
SRR30554315 /root/qiime2/input/SRR30554315_1.fastq.gz /root/qiime2/input/SRR30554315_2.fastq.gz
SRR30554318 /root/qiime2/input/SRR30554318_1.fastq.gz /root/qiime2/input/SRR30554318_2.fastq.gz
SRR30554319 /root/qiime2/input/SRR30554319_1.fastq.gz /root/qiime2/input/SRR30554319_2.fastq.gz
SRR30554316 /root/qiime2/input/SRR30554316_1.fastq.gz /root/qiime2/input/SRR30554316_2.fastq.gz
SRR30554324 /root/qiime2/input/SRR30554324_1.fastq.gz /root/qiime2/input/SRR30554324_2.fastq.gz
SRR30554325 /root/qiime2/input/SRR30554325_1.fastq.gz /root/qiime2/input/SRR30554325_2.fastq.gz
SRR30554317 /root/qiime2/input/SRR30554317_1.fastq.gz /root/qiime2/input/SRR30554317_2.fastq.gz
SRR30554328 /root/qiime2/input/SRR30554328_1.fastq.gz /root/qiime2/input/SRR30554328_2.fastq.gz
SRR30554323 /root/qiime2/input/SRR30554323_1.fastq.gz /root/qiime2/input/SRR30554323_2.fastq.gz
SRR30554337 /root/qiime2/input/SRR30554337_1.fastq.gz /root/qiime2/input/SRR30554337_2.fastq.gz
SRR30554339 /root/qiime2/input/SRR30554339_1.fastq.gz /root/qiime2/input/SRR30554339_2.fastq.gz
SRR30554326 /root/qiime2/input/SRR30554326_1.fastq.gz /root/qiime2/input/SRR30554326_2.fastq.gz
SRR30554341 /root/qiime2/input/SRR30554341_1.fastq.gz /root/qiime2/input/SRR30554341_2.fastq.gz
SRR30554342 /root/qiime2/input/SRR30554342_1.fastq.gz /root/qiime2/input/SRR30554342_2.fastq.gz
SRR30554327 /root/qiime2/input/SRR30554327_1.fastq.gz /root/qiime2/input/SRR30554327_2.fastq.gz
SRR30554343 /root/qiime2/input/SRR30554343_1.fastq.gz /root/qiime2/input/SRR30554343_2.fastq.gz
SRR30554355 /root/qiime2/input/SRR30554355_1.fastq.gz /root/qiime2/input/SRR30554355_2.fastq.gz
SRR30554330 /root/qiime2/input/SRR30554330_1.fastq.gz /root/qiime2/input/SRR30554330_2.fastq.gz
SRR30554357 /root/qiime2/input/SRR30554357_1.fastq.gz /root/qiime2/input/SRR30554357_2.fastq.gz
SRR30554361 /root/qiime2/input/SRR30554361_1.fastq.gz /root/qiime2/input/SRR30554361_2.fastq.gz
SRR30554331 /root/qiime2/input/SRR30554331_1.fastq.gz /root/qiime2/input/SRR30554331_2.fastq.gz
SRR30554362 /root/qiime2/input/SRR30554362_1.fastq.gz /root/qiime2/input/SRR30554362_2.fastq.gz
SRR30554364 /root/qiime2/input/SRR30554364_1.fastq.gz /root/qiime2/input/SRR30554364_2.fastq.gz
SRR30554332 /root/qiime2/input/SRR30554332_1.fastq.gz /root/qiime2/input/SRR30554332_2.fastq.gz
SRR30554365 /root/qiime2/input/SRR30554365_1.fastq.gz /root/qiime2/input/SRR30554365_2.fastq.gz
SRR30554369 /root/qiime2/input/SRR30554369_1.fastq.gz /root/qiime2/input/SRR30554369_2.fastq.gz
SRR30554333 /root/qiime2/input/SRR30554333_1.fastq.gz /root/qiime2/input/SRR30554333_2.fastq.gz
SRR30554370 /root/qiime2/input/SRR30554370_1.fastq.gz /root/qiime2/input/SRR30554370_2.fastq.gz
SRR30554376 /root/qiime2/input/SRR30554376_1.fastq.gz /root/qiime2/input/SRR30554376_2.fastq.gz
SRR30554334 /root/qiime2/input/SRR30554334_1.fastq.gz /root/qiime2/input/SRR30554334_2.fastq.gz
SRR30554377 /root/qiime2/input/SRR30554377_1.fastq.gz /root/qiime2/input/SRR30554377_2.fastq.gz
SRR30554381 /root/qiime2/input/SRR30554381_1.fastq.gz /root/qiime2/input/SRR30554381_2.fastq.gz
SRR30554345 /root/qiime2/input/SRR30554345_1.fastq.gz /root/qiime2/input/SRR30554345_2.fastq.gz
SRR30554384 /root/qiime2/input/SRR30554384_1.fastq.gz /root/qiime2/input/SRR30554384_2.fastq.gz
SRR30554346 /root/qiime2/input/SRR30554346_1.fastq.gz /root/qiime2/input/SRR30554346_2.fastq.gz
SRR30554348 /root/qiime2/input/SRR30554348_1.fastq.gz /root/qiime2/input/SRR30554348_2.fastq.gz
SRR30554349 /root/qiime2/input/SRR30554349_1.fastq.gz /root/qiime2/input/SRR30554349_2.fastq.gz
SRR30554350 /root/qiime2/input/SRR30554350_1.fastq.gz /root/qiime2/input/SRR30554350_2.fastq.gz
SRR30554263 /root/qiime2/input/SRR30554263_1.fastq.gz /root/qiime2/input/SRR30554263_2.fastq.gz
SRR30554268 /root/qiime2/input/SRR30554268_1.fastq.gz /root/qiime2/input/SRR30554268_2.fastq.gz
SRR30554352 /root/qiime2/input/SRR30554352_1.fastq.gz /root/qiime2/input/SRR30554352_2.fastq.gz
SRR30554274 /root/qiime2/input/SRR30554274_1.fastq.gz /root/qiime2/input/SRR30554274_2.fastq.gz
SRR30554275 /root/qiime2/input/SRR30554275_1.fastq.gz /root/qiime2/input/SRR30554275_2.fastq.gz
SRR30554354 /root/qiime2/input/SRR30554354_1.fastq.gz /root/qiime2/input/SRR30554354_2.fastq.gz
SRR30554276 /root/qiime2/input/SRR30554276_1.fastq.gz /root/qiime2/input/SRR30554276_2.fastq.gz
SRR30554284 /root/qiime2/input/SRR30554284_1.fastq.gz /root/qiime2/input/SRR30554284_2.fastq.gz
SRR30554287 /root/qiime2/input/SRR30554287_1.fastq.gz /root/qiime2/input/SRR30554287_2.fastq.gz
SRR30554356 /root/qiime2/input/SRR30554356_1.fastq.gz /root/qiime2/input/SRR30554356_2.fastq.gz
SRR30554289 /root/qiime2/input/SRR30554289_1.fastq.gz /root/qiime2/input/SRR30554289_2.fastq.gz
SRR30554290 /root/qiime2/input/SRR30554290_1.fastq.gz /root/qiime2/input/SRR30554290_2.fastq.gz
SRR30554360 /root/qiime2/input/SRR30554360_1.fastq.gz /root/qiime2/input/SRR30554360_2.fastq.gz
SRR30554291 /root/qiime2/input/SRR30554291_1.fastq.gz /root/qiime2/input/SRR30554291_2.fastq.gz
SRR30554294 /root/qiime2/input/SRR30554294_1.fastq.gz /root/qiime2/input/SRR30554294_2.fastq.gz
SRR30554363 /root/qiime2/input/SRR30554363_1.fastq.gz /root/qiime2/input/SRR30554363_2.fastq.gz
SRR30554298 /root/qiime2/input/SRR30554298_1.fastq.gz /root/qiime2/input/SRR30554298_2.fastq.gz
SRR30554301 /root/qiime2/input/SRR30554301_1.fastq.gz /root/qiime2/input/SRR30554301_2.fastq.gz
SRR30554372 /root/qiime2/input/SRR30554372_1.fastq.gz /root/qiime2/input/SRR30554372_2.fastq.gz
SRR30554302 /root/qiime2/input/SRR30554302_1.fastq.gz /root/qiime2/input/SRR30554302_2.fastq.gz
SRR30554305 /root/qiime2/input/SRR30554305_1.fastq.gz /root/qiime2/input/SRR30554305_2.fastq.gz
SRR30554373 /root/qiime2/input/SRR30554373_1.fastq.gz /root/qiime2/input/SRR30554373_2.fastq.gz
SRR30554310 /root/qiime2/input/SRR30554310_1.fastq.gz /root/qiime2/input/SRR30554310_2.fastq.gz
SRR30554312 /root/qiime2/input/SRR30554312_1.fastq.gz /root/qiime2/input/SRR30554312_2.fastq.gz
SRR30554374 /root/qiime2/input/SRR30554374_1.fastq.gz /root/qiime2/input/SRR30554374_2.fastq.gz
SRR30554314 /root/qiime2/input/SRR30554314_1.fastq.gz /root/qiime2/input/SRR30554314_2.fastq.gz
SRR30554320 /root/qiime2/input/SRR30554320_1.fastq.gz /root/qiime2/input/SRR30554320_2.fastq.gz
SRR30554378 /root/qiime2/input/SRR30554378_1.fastq.gz /root/qiime2/input/SRR30554378_2.fastq.gz
SRR30554321 /root/qiime2/input/SRR30554321_1.fastq.gz /root/qiime2/input/SRR30554321_2.fastq.gz
SRR30554322 /root/qiime2/input/SRR30554322_1.fastq.gz /root/qiime2/input/SRR30554322_2.fastq.gz
SRR30554379 /root/qiime2/input/SRR30554379_1.fastq.gz /root/qiime2/input/SRR30554379_2.fastq.gz
SRR30554329 /root/qiime2/input/SRR30554329_1.fastq.gz /root/qiime2/input/SRR30554329_2.fastq.gz
SRR30554335 /root/qiime2/input/SRR30554335_1.fastq.gz /root/qiime2/input/SRR30554335_2.fastq.gz
SRR30554380 /root/qiime2/input/SRR30554380_1.fastq.gz /root/qiime2/input/SRR30554380_2.fastq.gz
SRR30554336 /root/qiime2/input/SRR30554336_1.fastq.gz /root/qiime2/input/SRR30554336_2.fastq.gz
SRR30554338 /root/qiime2/input/SRR30554338_1.fastq.gz /root/qiime2/input/SRR30554338_2.fastq.gz
SRR30554385 /root/qiime2/input/SRR30554385_1.fastq.gz /root/qiime2/input/SRR30554385_2.fastq.gz
SRR30554340 /root/qiime2/input/SRR30554340_1.fastq.gz /root/qiime2/input/SRR30554340_2.fastq.gz
SRR30554344 /root/qiime2/input/SRR30554344_1.fastq.gz /root/qiime2/input/SRR30554344_2.fastq.gz
SRR30554387 /root/qiime2/input/SRR30554387_1.fastq.gz /root/qiime2/input/SRR30554387_2.fastq.gz
SRR30554347 /root/qiime2/input/SRR30554347_1.fastq.gz /root/qiime2/input/SRR30554347_2.fastq.gz
SRR30554351 /root/qiime2/input/SRR30554351_1.fastq.gz /root/qiime2/input/SRR30554351_2.fastq.gz
SRR30554353 /root/qiime2/input/SRR30554353_1.fastq.gz /root/qiime2/input/SRR30554353_2.fastq.gz
SRR30554358 /root/qiime2/input/SRR30554358_1.fastq.gz /root/qiime2/input/SRR30554358_2.fastq.gz
SRR30554359 /root/qiime2/input/SRR30554359_1.fastq.gz /root/qiime2/input/SRR30554359_2.fastq.gz
SRR30554366 /root/qiime2/input/SRR30554366_1.fastq.gz /root/qiime2/input/SRR30554366_2.fastq.gz
SRR30554367 /root/qiime2/input/SRR30554367_1.fastq.gz /root/qiime2/input/SRR30554367_2.fastq.gz
SRR30554368 /root/qiime2/input/SRR30554368_1.fastq.gz /root/qiime2/input/SRR30554368_2.fastq.gz
SRR30554388 /root/qiime2/input/SRR30554388_1.fastq.gz /root/qiime2/input/SRR30554388_2.fastq.gz
SRR30554371 /root/qiime2/input/SRR30554371_1.fastq.gz /root/qiime2/input/SRR30554371_2.fastq.gz
SRR30554375 /root/qiime2/input/SRR30554375_1.fastq.gz /root/qiime2/input/SRR30554375_2.fastq.gz
SRR30554382 /root/qiime2/input/SRR30554382_1.fastq.gz /root/qiime2/input/SRR30554382_2.fastq.gz
SRR30554383 /root/qiime2/input/SRR30554383_1.fastq.gz /root/qiime2/input/SRR30554383_2.fastq.gz
SRR30554386 /root/qiime2/input/SRR30554386_1.fastq.gz /root/qiime2/input/SRR30554386_2.fastq.gz
3) Mapping 파일 제작하기
Mapping 파일은 각 샘플별 metadata 정보가 들어있습니다. Manifest와 마찬가지로, 샘플에 대한 열 이름은 sample-id 이며, 각 열마다 '탭'으로 구분합니다. 그외에도, Mapping 파일을 만드는 규칙은 🔗 열이름 규칙 을 참고해주세요.
sample-id Source Case
SRR30554265 Stool Patient
SRR30554270 Stool Healthy
SRR30554271 Stool Healthy
SRR30554273 Stool Healthy
SRR30554277 Stool Healthy
SRR30554264 Colon Patient
SRR30554266 Stool Patient
SRR30554278 Stool Patient
SRR30554267 Stool Patient
SRR30554269 Stool Healthy
SRR30554279 Stool Patient
SRR30554272 Stool Healthy
SRR30554280 Colon Patient
SRR30554281 Stool Patient
SRR30554282 Stool Patient
SRR30554283 Stool Patient
SRR30554292 Stool Patient
SRR30554285 Stool Patient
SRR30554286 Stool Patient
SRR30554293 Stool Patient
SRR30554288 Colon Patient
SRR30554296 Stool Patient
SRR30554295 Stool Patient
SRR30554297 Terminal ileum Patient
SRR30554300 Terminal ileum Patient
SRR30554299 Terminal ileum Patient
SRR30554303 Stool Patient
SRR30554304 Stool Patient
SRR30554309 Stool Patient
SRR30554306 Stool Patient
SRR30554307 Stool Patient
SRR30554311 Colon Patient
SRR30554308 Stool Patient
SRR30554313 Terminal ileum Patient
SRR30554315 Terminal ileum Patient
SRR30554318 Terminal ileum Patient
SRR30554319 Terminal ileum Patient
SRR30554316 Terminal ileum Patient
SRR30554324 Terminal ileum Patient
SRR30554325 Terminal ileum Patient
SRR30554317 Terminal ileum Patient
SRR30554328 Terminal ileum Patient
SRR30554323 Terminal ileum Patient
SRR30554337 Terminal ileum Patient
SRR30554339 Colon Patient
SRR30554326 Terminal ileum Patient
SRR30554341 Colon Patient
SRR30554342 Colon Patient
SRR30554327 Terminal ileum Patient
SRR30554343 Colon Patient
SRR30554355 Colon Patient
SRR30554330 Terminal ileum Patient
SRR30554357 Colon Patient
SRR30554361 Colon Patient
SRR30554331 Terminal ileum Patient
SRR30554362 Stool Healthy
SRR30554364 Stool Healthy
SRR30554332 Terminal ileum Patient
SRR30554365 Stool Healthy
SRR30554369 Stool Healthy
SRR30554333 Colon Patient
SRR30554370 Stool Healthy
SRR30554376 Stool Healthy
SRR30554334 Terminal ileum Patient
SRR30554377 Stool Healthy
SRR30554381 Stool Healthy
SRR30554345 Colon Patient
SRR30554384 Stool Healthy
SRR30554346 Colon Patient
SRR30554348 Colon Patient
SRR30554349 Colon Patient
SRR30554350 Colon Patient
SRR30554263 Stool Healthy
SRR30554268 Stool Patient
SRR30554352 Colon Patient
SRR30554274 Stool Healthy
SRR30554275 Stool Healthy
SRR30554354 Colon Patient
SRR30554276 Stool Healthy
SRR30554284 Stool Patient
SRR30554287 Stool Patient
SRR30554356 Colon Patient
SRR30554289 Stool Patient
SRR30554290 Stool Patient
SRR30554360 Stool Healthy
SRR30554291 Stool Patient
SRR30554294 Stool Patient
SRR30554363 Stool Healthy
SRR30554298 Terminal ileum Patient
SRR30554301 Colon Patient
SRR30554372 Colon Patient
SRR30554302 Stool Patient
SRR30554305 Stool Patient
SRR30554373 Stool Healthy
SRR30554310 Stool Patient
SRR30554312 Terminal ileum Patient
SRR30554374 Stool Healthy
SRR30554314 Terminal ileum Patient
SRR30554320 Terminal ileum Patient
SRR30554378 Stool Healthy
SRR30554321 Terminal ileum Patient
SRR30554322 Terminal ileum Patient
SRR30554379 Stool Healthy
SRR30554329 Terminal ileum Patient
SRR30554335 Terminal ileum Patient
SRR30554380 Stool Healthy
SRR30554336 Terminal ileum Patient
SRR30554338 Colon Patient
SRR30554385 Stool Healthy
SRR30554340 Colon Patient
SRR30554344 Terminal ileum Patient
SRR30554387 Colon Patient
SRR30554347 Colon Patient
SRR30554351 Colon Patient
SRR30554353 Colon Patient
SRR30554358 Stool Healthy
SRR30554359 Stool Healthy
SRR30554366 Stool Healthy
SRR30554367 Stool Healthy
SRR30554368 Stool Healthy
SRR30554388 Colon Patient
SRR30554371 Stool Healthy
SRR30554375 Stool Healthy
SRR30554382 Stool Healthy
SRR30554383 Stool Healthy
SRR30554386 Colon Patient
QIIME2 환경 설정 🛠️
분석하기에 앞서, QIIME2 환경을 설정해야 합니다. Conda로 새 환경을 생성하는 방법에 대한 내용은 아래 포스팅을 참고해주세요 :)
🔗 2025.03.03 - [생물정보/Metagenome] - [QIIME2] QIIME2 설치
[QIIME2] QIIME2 설치
* 리눅스 서버 환경이 필요합니다. Anaconda/Miniconda를 활용한 QIIME2 설치 🐍💻Anaconda 혹은 Miniconda는 Python 환경을 쉽게 관리할 수 있는 도구로, QIIME2 설치 시 가장 일반적으로 사용됩니다. Anaconda는 P
jeonyeong.tistory.com
QIIME2 환경을 생성하셨다면, 아래의 명령어를 통해 QIIME2 환경을 불러오세요.
(참고로 저의 qiime2 환경 이름은 qiime2-amplicon-2024.10 입니다.)
# conda activate [qiime2 환경 이름]
conda activate qiime2-amplicon-2024.10QIIME2 결과 파일 확인하기 👓
QIIME2 결과 파일들은 .qza 혹은 .qzv 포맷으로 결과들을 제공해줍니다.
.qza : 데이터 파일
.qzv : .qza 파일을 시각화한 파일
일반적으로 .qza파일을 바로 읽지 않고, .qzv 로 변환하여 데이터를 확인합니다. 데이터를 보기 위해서는 QIIME2 명령어를 활용하여 파일을 해제 할 수도 있지만, 🔗QIIME2 View 사이트에서 qzv 파일을 드래그 앤 드랍하여 확인 할 수 있습니다.
데이터 업로드하기☁️
1) Data Importing
먼저, raw data를 불러옵니다. 보통 Illumina 시퀀싱 데이터는 .fastq 형식으로 제공되며, QIIME2의 qza 포맷으로 변환해야 합니다.
(* 모든 결과 파일은 /root/qiime2/output 경로에 저장해둘게요.)
cd /root/qiime2/
mkdir -p output/
qiime tools import \
--type SampleData[PairedEndSequencesWithQuality] \
--input-path manifest.txt \
--output-path output/demux.qza \
--input-format PairedEndFastqManifestPhred33V22) Data visualization
업로드된 raw data를 시각화하여 결과를 확인할 수 있습니다.
qiime demux summarize \
--i-data output/demux.qza \
--o-visualization output/demux.qzvOutput
(파일을 다운로드 받아 🔗QIIME2 View 에서 확인해보세요.)
결과 해석하기 👩🏻💻
📂demux.qzv
1) Demultiplexed sequence counts summary
입력한 데이터는 paired-end 서열이라, forward reads, reverse reads 에 대한 통계량 정보가 제공됩니다.
입력한 샘플들 중 최소 / 중앙값 / 평균 / 최대 리드수와 전체 리드수에 대한 정보입니다.
그리고 이에 대한 히스토그램을 Demultiplexed sequence counts summary 테이블 아래에서 제공합니다.


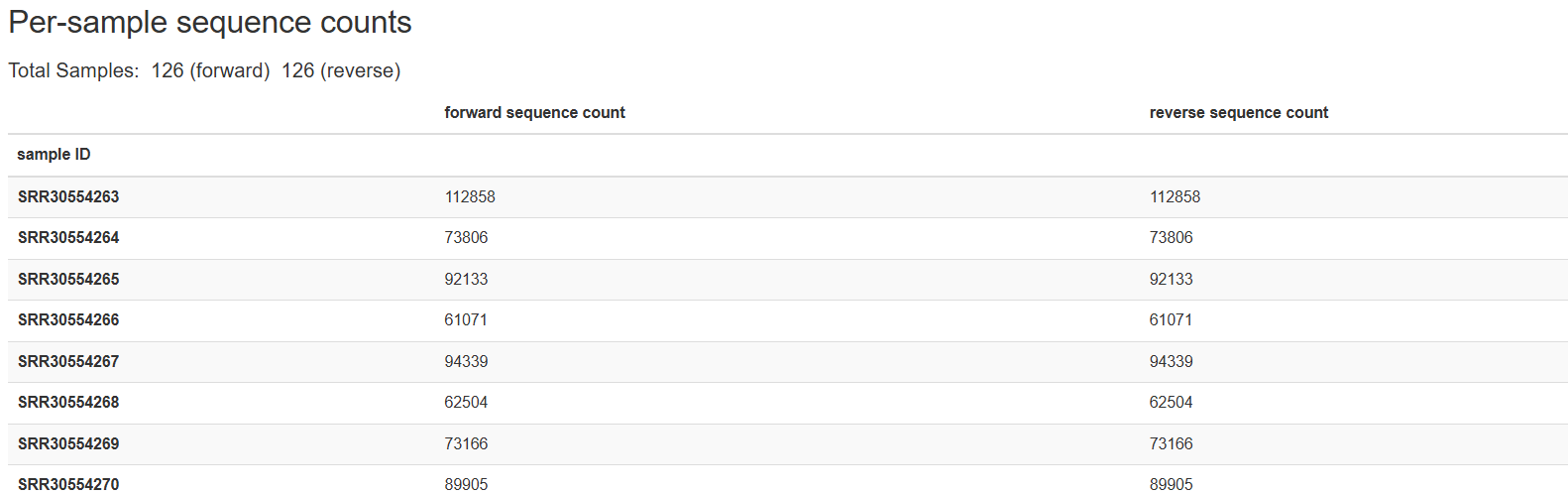
2) Per-sample sequence counts
입력한 샘플에 대한, 샘플별 forward / reverse 서열 수에 대한 테이블 입니다.
3) Interactive Quality Plot
Forward / Reverse 서열에 대한 Quality Plot을 제공합니다. 원하는 영역을 드래그하여 확대할 수 있습니다. 더블클릭하면 다시 원래대로 돌아올 수 있습니다.
참고로, 일반적으로 Illumina 플랫폼의 경우 서열 말단의 퀄리티가 떨어지는 양상을 보입니다.

마우스를 Quality Plot에 오버랩하면, 해당위치(bp)에 대한 통계량 정보를 아래의 테이블에서 확인 할 수 있습니다. 전체 테이블은 테이블 아래의Download forward/reverse parametric seven-number summaries as TSV 버튼을 클릭하여 TSV 포멧으로 파일을 다운로드 받을 수 있습니다.

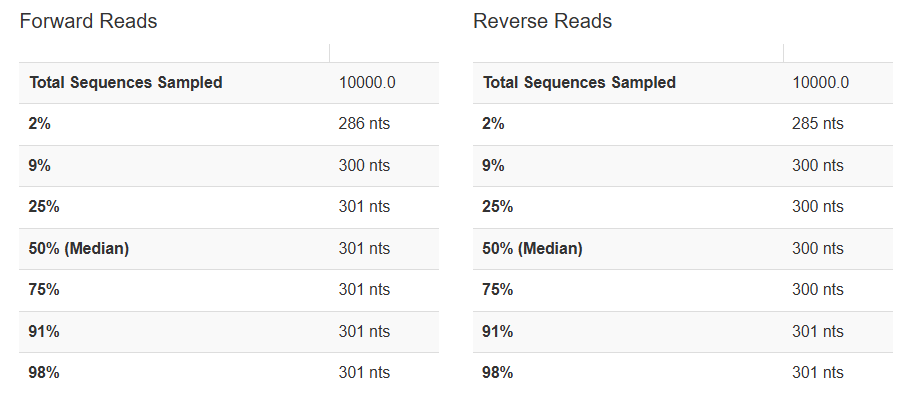
4) Demultiplexed sequence length summary
Forward / Reverse 서열의 길이에 대한 통계량 파일입니다.
🔗 링크
QIIME2 Moving picture tutorial : https://docs.qiime2.org/2024.10/tutorials/moving-pictures/
QIIME2 View : https://view.qiime2.org/
'생물정보 > Metagenome' 카테고리의 다른 글
| [QIIME2] QIIME2 실습 - (2) 데이터 전처리 (0) | 2025.03.17 |
|---|
